Biosensors modeling and simulations: a new way to discover drugs
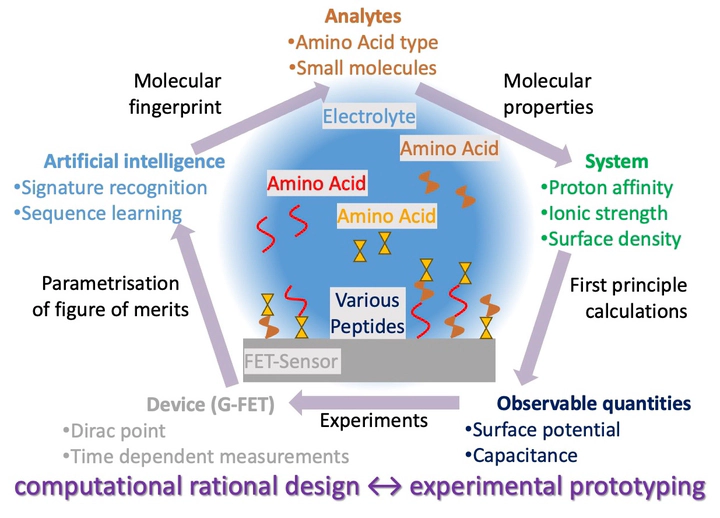
We are building a unique simulation framework that combines machine learning methods and device simulation techniques.
Our biosensor methodology is based on the combination of machine learning methods and ‘classical’ device simulations. We have developed a unique simulation framework that combines the Site-binding and Gouy-Chapman-Stern (GCS) analytical models, calibrated to the experimental data, with a machine learning (ML) approach. The ML model, which we call the BioTokens model, is based on a transformative network. As an input, the BioTokens model uses a combination of surface potential and capacitance obtained from GCS and a sequence of ’tokens’.
The output of the simulation tool is a sequence of amino acids or small oligopeptides, which can be compared with the surface potential and capacitance obtained from experiments or analytical simulations. The main strength of our approach is that we can predict the surface potential and capacitance of proteins and amino acids.
Selected publications:
Smoliak, N., Kumar, N. , Parreira, P. M. , Macdonald, C. and Georgiev, V. (2025) A hybrid analytical and machine learning model of BioFETs for the detection of peptides. IEEE Sensors Letters, 9(10), 4502604. doi: 10.1109/LSENS.2025.3606612
Kumar, N., Dixit, A., Kumar, P., Ansari, Md. H., Bagga, N., Gandhi, N., Kondekar, P.N., García, C. P. and Georgiev, V. (2025) Optimizing peptide detection using FET-based sensors: integrating non-linearities of surface functionalization. Solid-State Electronics, 229, 109161. doi: 10.1016/j.sse.2025.109161
Kumar, N. , Aleksandrov, P. , Gao, Y., Macdonald, C. , García, C. P. and Georgiev, V. (2024) Combinations of analytical and machine learning methods in a single simulation framework for amphoteric molecules detection. IEEE Sensors Letters, 8(7), 1501004. doi: 10.1109/LSENS.2024.3408101
Kumar, N. , Dhar, R. P., El Maiss, J., Georgiev, V. and García, C. P. (2024) Discovery of amphoteric fingerprints of amino acids with field-effect transistors. IEEE Access, doi: 10.1109/ACCESS.2024.3411168
Dhar, R., Kumar, N. , Pascual Garcia, C. and Georgiev, V. (2022) Assessing the effect of scaling high-aspect-ratio ISFET with physical model interface for nano-biosensing application. Solid-State Electronics, 195, 108374. doi: 10.1016/j.sse.2022.108374
Kumar, N. , Pascual García, C., Dixit, A., Rezaei, A. and Georgiev, V. (2023) Charge dynamics of amino acids fingerprints and the effect of density on FinFET-based electrolyte-gated sensor. Solid-State Electronics, 210, 108789. doi: 10.1016/j.sse.2023.108789
Kumar, N. , Dhar, R. P. S., Pascual Garcia, C. and Georgiev, V. (2023) A novel computational framework for simulations of bio-field effect transistors. ECS Transactions, 111(1), pp. 249-260. doi: 10.1149/11101.0249ecst